Phytophthora ramorum
P. ramorum Werres, De Cock, & Man in’t Veld (2001) appears to be an exotic species introduced from an unknown origin to Europe and western N. America in the mid-1990s, likely on nursery plants (Mascheretti et al. 2008). In California and southwest Oregon, the pathogen spread to native oak and tanoak forests, where it causes lethal stem cankers (sudden oak death). Between 30-45 million trees have been killed. In Europe, where nurseries in over 20 countries reported P. ramorum, the disease on trees was limited to isolated stands of beech and oak associated with infected rhododendrons. In 2009, the pathogen began causing widespread mortality (sudden larch death) on Japanese larch timber plantations in the UK (Brasier & Webber 2010). International, national, state and county quarantines are in place to prevent the further spread of disease with nursery plants and to contain the infestation in forests. P. ramorum is unusual among forest Phytophthora species for its wide host range and infection of aerial plant parts.
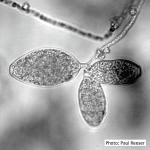
Sporangia (average 46–65 µm by 21–28 µm) formed on agar and in water, ellipsoid, spindle-shaped, or elongate-ovoid, semi-papillate, caducous with short pedicel (< 5 µm), produced singly or in clusters of 2–12 on sympodially branched sporangiophores. Chlamydospores (20–91 µm; average 46-60 µm) formed abundantly in agar, globose, mostly thin-walled, terminal or intercalary. Heterothallic, oogonia not seen in single culture (Werres et al. 2001).
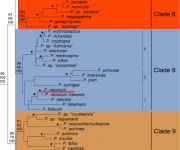
P. ramorum is placed in clade 8c, with P. lateralis as its closest relative (Blair et al., 2008). Populations in North America and Europe are clonal and belong to four lineages according to several molecular markers (Grunwald et al., 2008; Van Poucke et al. 2012; Gagnon et al. 2014).
Table 1. Characteristics of clonal lineages of P. ramorum (adapted from Grünwald et al. 2009 and others)
|
Clonal lineage |
Distribution |
Habitat |
Mating type |
|
EU1 |
Europe, N. America |
Nurseries, gardens, forest plantations, forests |
A1* |
|
EU2 |
UK (N. Ireland, SW Scotland) |
Forest plantations |
A1 |
|
NA1 |
N. America |
Forests, nurseries |
A2 |
|
NA2 |
N. America |
Nurseries |
A2 |
*Most EU1 isolates are mating type A1 but several isolates from Belgium are mating type A2
Evidence suggests that clonal lineages descended from a sexually recombining population but have been reproductively isolated for a long time (Goss et al. 2009). Oospores have not been observed under natural conditions despite the concurrent presence of both mating types in nurseries and Oregon forests. A 7X draft whole-genome sequence (Tyler et al. 2006) and mitochondrial sequence (Martin et al., 2007) of P. ramorum are available.
Phylogenetic tree from http://www.phytophthoradb.org/ramorum (Blair et al 2008).
Temperature optimum ca. 20°C, min. ca. 2°C, max. ca. 26–30°C. Growth at 20°C on V8 agar 2.2 mm/d, on cornmeal agar-β sitosterol 1.9 mm/d, on cornmeal agar with pimaricin, ampicillin, and rifampicin (PAR) 0.7 mm/d, on potato dextrose agar 1.2 mm/d.
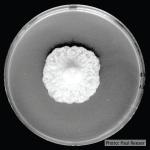
Colony morphology and hyphal characteristics on isolation plates are distinctive. The abundance of large chlamydospores in combination with clusters of semi-papillate, caducous, ellipsoid sporangia formed on the surface of the agar may be considered definitive. Be aware that these features may be seen independently in other Phytophthora species growing on isolation plates.
The searchable web-based database Phytophthora-ID is useful for rapid identification of Phytophthora species based on sequencing of the ITS or Cox spacer regions, followed by BLAST searching the database. Phytophthora-ID maintains a database of sequences that is selective for sequence accessions that come from trusted sources including published, peer-reviewed studies whenever possible.
For more information about Phytophthora ramorum, visit our Disease, Education and Management materials, and Reference sections.